Difference between revisions of "NiftyReg Boundary Shift Integral Tutorial"
| Line 1: | Line 1: | ||
| − | Starting from two longitudinal images from the same subject and associated segmentations of region of interest, one can use the Boundary Shift Integral (BSI) to assess the atrophy of the region of interest over time. | + | Starting from two longitudinal images from the same subject and associated segmentations of region of interest, one can use the Boundary Shift Integral (BSI) to assess the atrophy of the region of interest over time. The computation of BSI requires the input images to be co-register. This tutorial focuses only on this co-registration step. |
| − | The images used in this tutorial are extracted from the MIRIAD database. | + | The images used in this tutorial are extracted from the [http://dx.doi.org/10.1016/j.neuroimage.2012.12.044 MIRIAD database]. The full database can be downloaded from [http://www.ucl.ac.uk/drc/research/miriad-scan-database this page] |
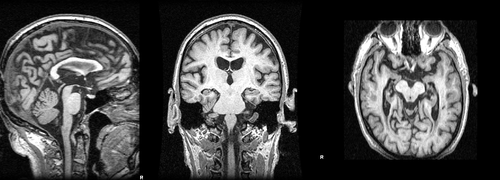
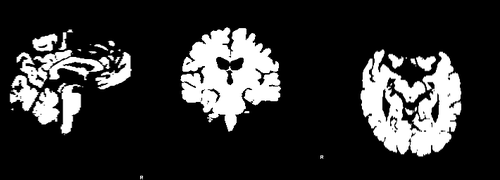
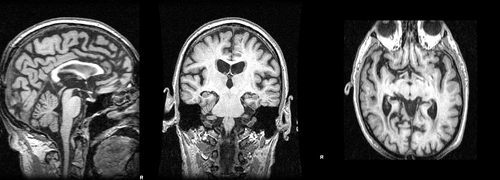
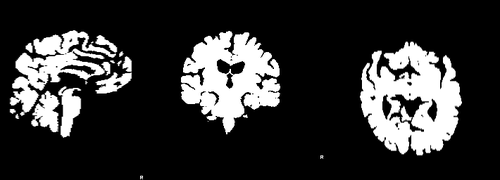
Images 188_A.nii.gz and brain_188_A.nii.gz are a T1-weighted MRI acquired at baseline and its associted full brain segmentation. Images 188_F.nii.gz and brain_188_F.nii.gz are the equivalent images for the follow-up acquisition. | Images 188_A.nii.gz and brain_188_A.nii.gz are a T1-weighted MRI acquired at baseline and its associted full brain segmentation. Images 188_F.nii.gz and brain_188_F.nii.gz are the equivalent images for the follow-up acquisition. | ||
| Line 33: | Line 33: | ||
::<code>-res brain_188_F_midspace.nii.gz -inter 0</code> | ::<code>-res brain_188_F_midspace.nii.gz -inter 0</code> | ||
| − | Using the 4 input images resampled into the mid-space, we can now run the BSI. One implementation can be found on this [http://sourceforge.net/projects/bsintegral sourceforge page]. | + | Using the 4 input images resampled into the mid-space, we can now run the BSI. In practice, a [http://dx.doi.org/10.1016/j.neuroimage.2004.04.030 differential bias field correction] is usually performed on those images. |
| + | One implementation of BSI can be found on this [http://sourceforge.net/projects/bsintegral sourceforge page]. | ||
Latest revision as of 13:08, 11 March 2015
Starting from two longitudinal images from the same subject and associated segmentations of region of interest, one can use the Boundary Shift Integral (BSI) to assess the atrophy of the region of interest over time. The computation of BSI requires the input images to be co-register. This tutorial focuses only on this co-registration step.
The images used in this tutorial are extracted from the MIRIAD database. The full database can be downloaded from this page
Images 188_A.nii.gz and brain_188_A.nii.gz are a T1-weighted MRI acquired at baseline and its associted full brain segmentation. Images 188_F.nii.gz and brain_188_F.nii.gz are the equivalent images for the follow-up acquisition.
In order to run BSI, we first need to register the baseline and followup image.
reg_aladin -ref 188_A.nii.gz -flo 188_F.nii.gz \
-rmask dil_brain_188_A.nii.gz -fmask dil_brain_188_F.nii.gz \-res ref_A_flo_F_aff_res.nii.gz -aff ref_A_flo_F_aff_mat.txt
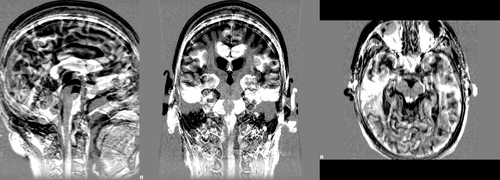
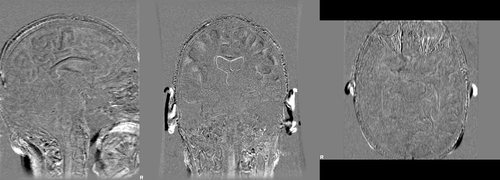
where images dil_brain_188_A.nii.gz and dil_brain_188_F.nii.gz are dilated version of the brain masks of the baseline and followup images respectively. The following images show difference images without registration and with registration respectively
Both input images are then resampled into a mid-space to ensure that no interpolation bias affect the directionality of the BSI estimation. The affine transformations to mid-space are computed using the following commands:
reg_transform -half ref_A_flo_F_aff_matrix.txt ref_A_flo_F_half_aff_matrix.txtreg_transform -invAff ref_A_flo_F_half_aff_matrix.txt ref_F_flo_A_half_aff_matrix.txt
Baseline and followup images are then resampled using the generated transformation:
reg_resample -ref 188_A.nii.gz -flo 188_A.nii.gz -trans ref_F_flo_A_half_aff_matrix.txt \
-res 188_A_midspace.nii.gz
reg_resample -ref 188_A.nii.gz -flo 188_F.nii.gz -trans ref_A_flo_F_half_aff_matrix.txt \
-res 188_F_midspace.nii.gz
The associated brain segmentations are resampled to the same space using nearest-neighbour interpolation:
reg_resample -ref 188_A.nii.gz -flo brain_188_A.nii.gz -trans ref_F_flo_A_half_aff_matrix.txt \
-res brain_188_A_midspace.nii.gz -inter 0
reg_resample -ref 188_A.nii.gz -flo brain_188_F.nii.gz -trans ref_A_flo_F_half_aff_matrix.txt \
-res brain_188_F_midspace.nii.gz -inter 0
Using the 4 input images resampled into the mid-space, we can now run the BSI. In practice, a differential bias field correction is usually performed on those images. One implementation of BSI can be found on this sourceforge page.